Mutation Designer - Q5®-Like SDM primer design
Our motivation
We are a team of biochemists & molecular biologists, and we found ourselves frequently designing primers for performing several PCR-based whole-plasmid Site-Directed Mutagenesis (SDM) experiments. Designing of SDM primers requires inspection of the codon table and regular monitoring of the dynamically changing primer properties as the primer is being designed. To make our lives a little easier, we wrote Mutation Designer to automate the primer design workflow we currently use in our lab. We hope you'll find Mutation Designer useful.
General information
Using this tool you can generate SDM primers where the Forward primer carries the mutation and there is no overlap among the forward and reverse prmers. The following schematic illustrates the primer design strategy. Cloning strategies like this generally require 5'-phosphorylation (using a kinase, or synthetic oligos with 5'-Pho makes these easy).
As show in the following figure, the desired mutation is introduced by the forward primer initiating replication in the sense strand of the DNA, and the reverse primer starts on the antisense strand in the reverse direction.
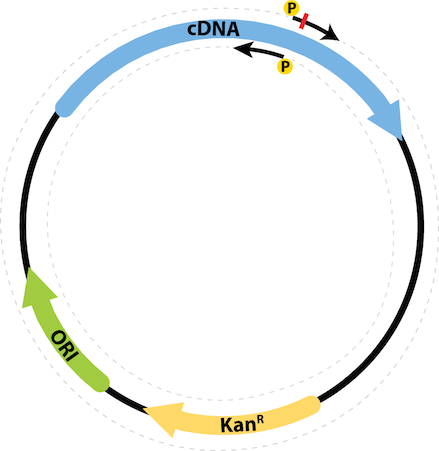
Subsequently, the PCR reaction is treated with DpnI restriction enzyme (which cuts methylated DNA, and spares the newly synthesized PCR amplicons) to digest the template DNA and the plasmid is circularized by using the a DNA ligase since the primers already have 5' Phosphate groups.
Mutation designer requires following inputs
- DNA Sequence : A text file with the target DNA sequence as a continuous string of characters.
DNA can be either in upper case, Lower case or even mixed case. DNA can be in several lines (e.g. if it is from a FASTA file), just remember to remove the header from fasta files (i.e. the first line starting with ‘>’). An example file can be downloaded by clicking “Example DNA” near the upload box.
xxxxxxxxxx
1
1
atgaagccggcgacaggactttgggtctgggtgagccttctcgtggcggcggggaccgtccagcccagcgattctcagtcagtgtgtgcaggaacggagaataaactgagctctctctctgacctggaacagcagtaccgagccttgcgcaagtactatgaaaactgt
- List of desired mutations : A text file listing the desired amino acid changes. Each amino acid change should be listed in a new line. An example file can be downloaded by clicking “Example list of mutations” near the upload box.
xxxxxxxxxx
5
1
A123T
2
V234Y
3
P456S
4
...
5
... continued
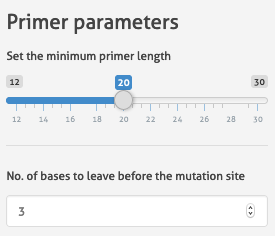
- Minimum length of the primers: User can also use the slider to adjust (figure below) the minimum desired length of primer. There no “right” values, a lot depends on your primer binding site (GC content, number of mismatches), your polymerase, your PCR conditions. Adjust it to what you use
- Number of bases preceeding the mutated nucleotide: Mutagenesis on the 5' end of the primer can lead to deletions instead of incorporation of the DNA alteration due to issues with primer annealing. Therefore, it is a standard practice to incorporate the mutated base as a 3-5 nucleotide in our lab. We have had good success with the strategy, but we're aware that other practices exist therefore we give the users freedom to set the number of preceeding bases they prefer to have.
Mutation Designer uses nearest neighbor thermodynamics to calculate annealing temperatures which is based on the method described in, Allawi & SantaLucia Jr, Nucleic Acids Res(1998) PMID:9592156.
Output of Mutation Designer
Mutation Designer accepts the above mentioned inputs and predicts primers for a whole-plasmid Site Directed Mutagenesis. Mutation Designer takes these into account the user inputs and attempts to design the "best possible" primer pairs depending on the local genetic context:
-
Primers creating the desired mutation with minimum number of nucleotide changes.
- In the event there are alternative ways to create a point mutation with the same number of nucleotide changes, Mutation designer lists all such possibilities.
-
Primers starting with a G or C at 5' end.
-
Primers that are at least 18nt long (user can change this threshold).
-
Primer pairs with a difference of at most 5°C in their annealing temperatures.
-
Primer pairs with a 40 to 60% GC content.
-
Primer pairs with GC content within 15% of each other.
Mutation Designer displays the output to the user on the wepbage on clicking the "Display Results" button.
Although, we strongly recommend clicking the "Download Results" button to obtain a TSV file with a table containing complete information about the predicted primers, including warnings (if any).
Warning Flags of Mutation Designer
There are four warning flags in the output table. Here is how to interpret them:
- Leading_AT_warning is "Y" if the Forward Primer doesn't start with G or C.
- GC_warning_FW is "Y" if the GC content for FW primer is < 40% or >60%.
- GC_warning_REV is "Y" if the GC content for REV primer is < 40% or >60%.
- GC_Diff_Warning is "Y" if the difference in the GC content of the FW and REV primers's is >15%.
In case of primers with warnings, we recommend a close inspection of the nucleotide landscape around the mutation site, and if possible, a re-designing of the primer sequences by altering the input parameters in Mutation Designer or a complete manual re-design (if the results from Mutation Designer are unsatisfactory).
Caveats & Disclaimer:
-
Number of mismatches in the mutagenic primer are not accounted for during the standard Tm calculations.
-
The Tm values are not tailored towards any particular polymerase or protocol. They are merely indicative. Please follow the directions of your PCR kit to calculate the correct annealing temperatures.
-
Like with most PCRs optimization of PCR conditions (Annealing temp, extension time, number of cycles, etc.) might be necessary to get desired results.
-
We are not affiliated with nor do we endorse QuickChange®, Q5®, or any of their competing products and their parent companies. These two products are widely used in their subtype of SDM protocols, therefore we used those names to give you, the user, a quick idea about the differences between our two variants of Mutation Designer.
Points to consider before ordering the oligos
-
Ensure that the forward primer(s) creates your desired amino acid change(s) and the reverse primer matches the anti-sense strand of your template to create a whole plasmid mutagenesis.
-
Check for potential off-target annealing sites for these primers in your host organism by using Primer Blast from NCBI.
-
Check that the primers do not have self-complimentarity and that the primers are not making any secondary structures using the Oligo Analyzer tool by IDT.
-
Remember to order 5’-Phospho oligonucleotides for convenience during ligation. But remember that even regular oligos can be phosphorylated using a polynucleotide kinase, e.g. the T4 Polynucleotide Kinase.
Privacy
Being researchers ourselves, we understand the confidentiality of your data, therefore Mutation designer deletes your input files as soon as the execution session ends.
References
- Hatim T. Allawi, John SantaLucia, Jr, Thermodynamics of internal C·T mismatches in DNA, Nucleic Acids Research, Volume 26, Issue 11, 1 June 1998, Pages 2694–2701, https://doi.org/10.1093/nar/26.11.2694
