Motivation
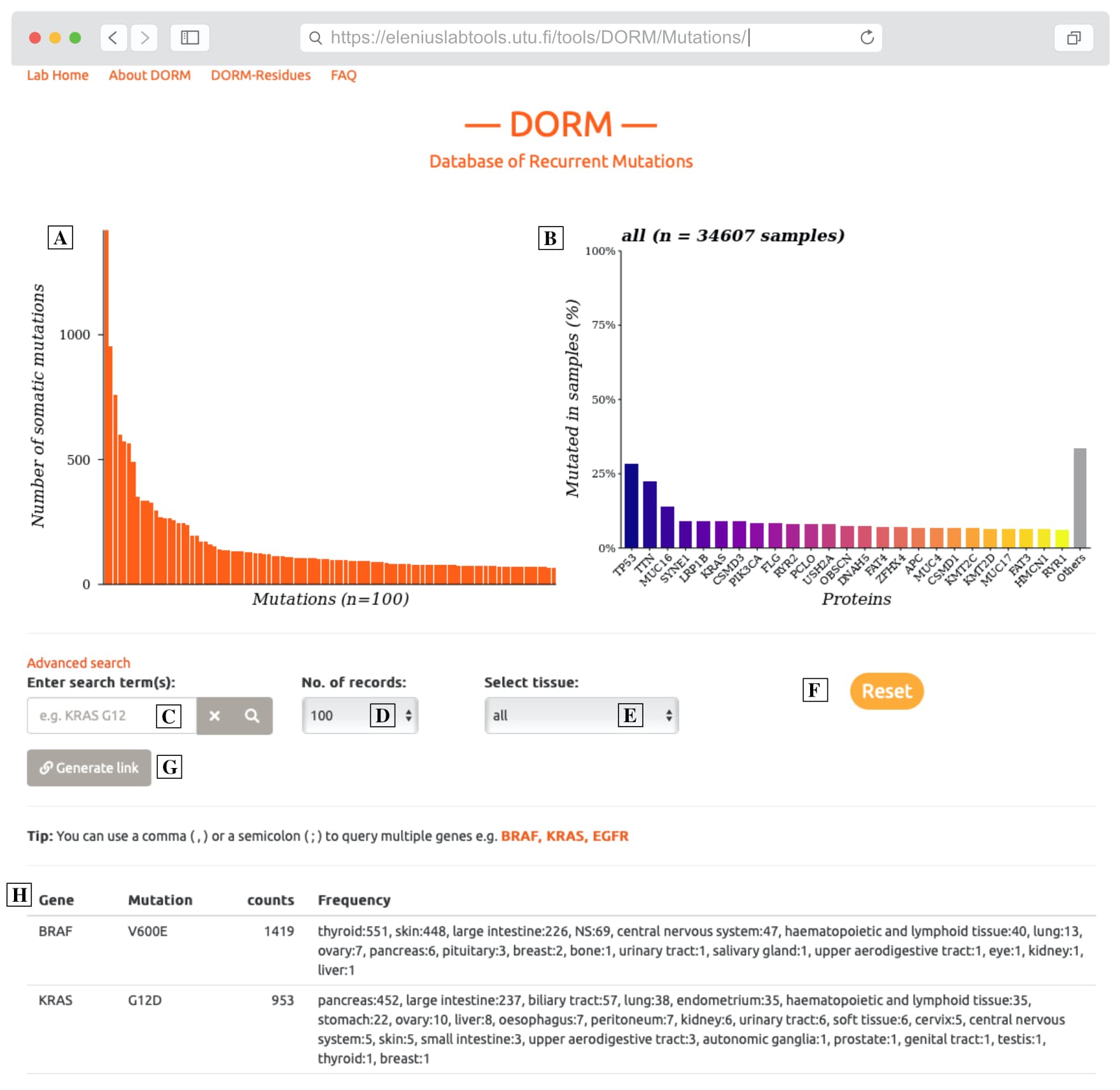
In our lab, we regularly find ourselves looking up various somatic mutations in the context of human cancers and retreive their frequency of occurance in human cancer samples. Therefore, we made this simple tool and dubbed it "DORM". We process the COSMIC data releases and present our processed database here which display the statistics about recurrent cancer-associated mutations that are identified from genome-wide screens (i.e., a mutation e.g. KRAS G12C, is listed in DORM if it has a tissue-agnostic population frequency > 1 ).
Our Goal
We aimed at developing a fast and lightweight web-tool to give a quick-and-easy peek into COSMIC dataset so a user can check if their particular mutation or mutations in their favorite protein(s) is common or rare occurence in cancer samples.
About the database
DORM is made available in two variants to answer two related, but slightly different questions. The user can also browse or restrict the search space to 38 tissue types e.g. pancreas, skin, lung, etc.
1. Individual Mutations
Here you can find the
frequency of the recurrent mutations listed as exact amino acid
changes. You can explore the the
database with search terms like: 'EGFR,
KRAS, BRAF' , 'EGFR L858R'
, and even perform advanced
searches using regular expressions.
2. Grouped by Residue
This database lists the frequency of the recurrent mutations grouped
by amino acid residues e.g. KRAS G12C/V/D are all grouped as KRAS G12. You can explore the
the
database with search terms like:
'EGFR, KRAS, BRAF' , 'KRAS Q' , and even perform advanced searches using
regular expressions.
How to use the database?